Using alternative buffer for DNA genomic isolation in forest trees
Downloads
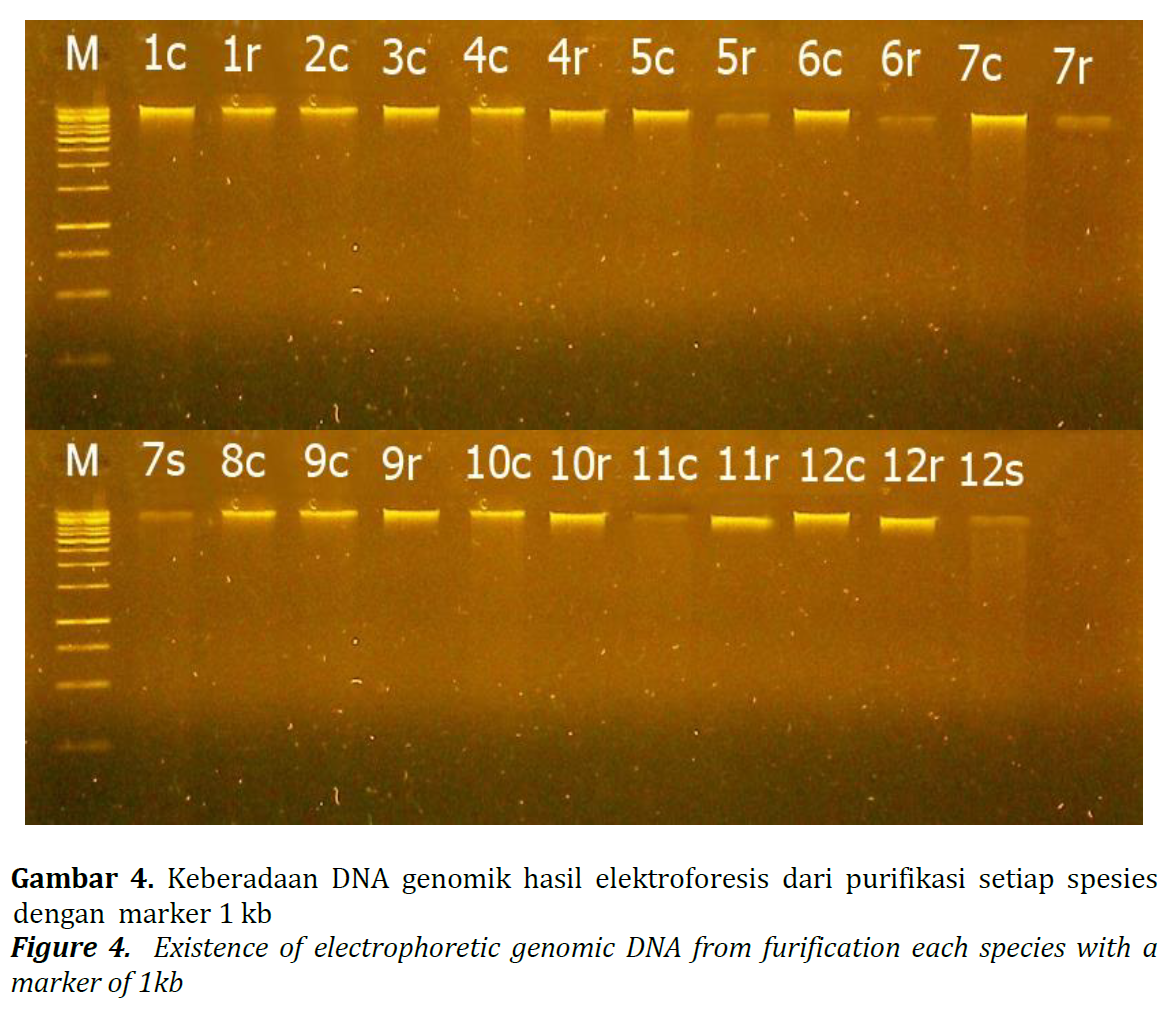
DNA optimization procedures can be carried out on the type of buffer used during extraction or physical handling techniques in separating genomic DNA from other compounds. The research using Three types of buffer: 1. CTAB, 2. Detergents containing Alkyl Benzene Sulfonate (ABS) surfactants and Three Detergents containing Sodium Laureth Sulphate (SLS) surfactants. This study aims to obtain an optimal DNA isolation method to produce genomic DNA of good quality and sufficient quantity so that it can be used for genetic diversity analysis in forest trees and to determine the optimal alternative buffer for CTAB to facilitate DNA isolation in remote areas, which generally hard to get CTAB. The parameters measured in this study were the presence of DNA and DNA concentration. The results showed that DNA isolation of 12 species of forest trees was successfully carried out using CTAB buffer and Detergent containing ABS surfactants by visualizing the genomic DNA bands from the results of the electrophoresis and Nanodrop spectrophotometer meanwhile Detergent containing SLS surfactants buffer was not successful in DNA isolation. The highest DNA quantity (DNA concentration) was found in 19 samples using CTAB buffer and the detergent containing ABS surfactants buffer with a concentration of 1403,8 - 3412,7 ng/μl. The conclusion of this study was CTAB buffer and the Detergent containing ABS surfactants can also be used as an alternative to a simple buffer for DNA isolation experiments.
Afshar, M., Mansour, R., Mohammad, H. F., & Seyyed, F. (2018). Comparative analysis and innovation of a simple and rapid method for high-quality RNA and DNA extraction of kiwifruit. MethodsX, 5, 352–361.
Apriyani, N. (2017). Penurunan kadar surfaktan dan sulfat dalam limbah laundry. Media Ilmiah Teknik Lingkungan, 2(1), 37–44.
Ari, P., Simanjuntak, P., & Suwarno, T. (2019). Pengaruh metoda ekstraksi simplisia multi herbal dan multi ekstrak daun sukun, seledri dan daun salam terhadap aktivitas antikolesterol secara in-vitro. Medika Tadulako, Jurnal Ilmiah Kedokteran, 6(2), 78–87.
Corkill, G., & Rapley, R. (2008). The manipulation of nucleic acids basic tools and techniques. In J. Walker (Ed.), Molecular Biomethods Handbook: Second Edition. Humana Press.
Couch, J. A., & Fritz, P. J. (1990). Isolation of DNA from plants high in polyphenolics. Plant Molecular Biology Reporter, 8, 8–12.
Doyle, J. ., & Doyle, J. . (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19(1), 11–15.
Hairuddin, R. (2013). Isolasi DNA dan amplifikasi (PCR) genom DNA kopi (Coffea Sp) melalui proses elektroforesis gel poliakrilamid. Dinamika, 4(1), 43–48.
Howell, S. H. (2014). Molecular biology. In S. H. Howell (Ed.), Molecular Biology (1st ed.). Springer-Verlag New York.
Langga, I. F., Restu, M., & Kuswinanti, T. (2012). Optimalisasi suhu dan lama inkubasi dalam ekstraksi DNA tanaman bitti (Vitex Cofassus Reinw) serta analisis keragaman genetik dengan teknik RAPD-PCR. J Sains & Teknologi, 12(3), 265–276.
Nugroho, K., Terryana, R. T., Rijzaani, H., & Lestari, P. (2016). Metode ekstraksi DNA pada Jatropha spp. tanpa menggunakan nitrogen cair/DNA extraction method of Jatropha spp. Without liquid nitrogen. Jurnal Penelitian Tanaman Industri, 22(4), 159–166.
Nurkamila, U. S., & Pharmawati, M. (2014). Ekstraksi DNA dari herbarium anggrek. SIMBIOSIS Journal of Biological Sciences, 2(1), 135–146.
Restu, M., Mukrimin, M., & Gusmiaty, G. (2012). Optimalisasi teknik ekstraksi dan isolasi DNA tanaman suren (Toona Sureni Merr.) untuk analisis keragaman genetik berdasarkan Random Amplified Polymorphic DNA (RAPD). Jurnal Natur Indonesia, 14(1), 136–142.
Robinson, T. (1974). Metabolism and function of alkaloids in plants. Science, 184(4135), 430–435
Sahoo, R. K., Ansari, M. W., Pradhan, M., Dangar, T. K., Mohanty, S., & Tuteja, N. (2014). Phenotypic and molecular characterization of native Azospirillum strains from rice fields to improve crop productivity. Protoplasma, 251, 943–953.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). In: Molecular cloning: A laboratory manual. In J. Sambrook (Ed.), Cold Spring Harbor (2nd ed.). Cold Spring Harbor Laboratory Press.
Syafaruddin, Randriani, E., & Santoso, T. J. (2011). Efektivitas dan efisiensi teknik isolasi dan purifikasi DNA pada jambu mete. Jurnal Tanaman Industri dan Penyegar, 2(2), 151-160.
Varma, A., Padh, H., & Shrivastava, N. (2007). Plant genomic DNA isolation: an art or a science. Biotechnology Journal, 2(3), 386–392.
Yi, S., Jin, W., Yuan, Y., & Fang, Y. (2018). An optimized CTAB method for genomic DNA extraction from freshly-picked pinnae of Fern, Adiantum capillus-veneris L. Bio-protocol, 8(13), e2906.
Yulianti, E. (2006). Pengembangan teknik isolasi DNA tumbuhan menggunakan detergen komersial. Seminar Nasional MIPA (Penelitian, Pendidikan, dan Penerapan MIPA serta Peranannya dalam Peningkatan Keprofesionalan Pendidik dan Tenaga Kependidikan) pada tanggal 1 Agustus 2006, 71–85, diselenggarakan oleh Fakultas Matematika dan Ilmu Pengetahuan Alam UNY, Yogyakarta.




